The CATH Database
Introduction
The CATH database is a hierarchical domain classification of protein structures in the Protein Data Bank (PDB, Berman et al. 2003). Only crystal structures solved to resolution better than 4.0 angstroms are considered, together with NMR structures. All non-proteins, models, and structures with greater than 30% “C-alpha only” are excluded from CATH. This filtering of the PDB is performed using the SIFT protocol (Michie et al., 1996). Protein structures are classified using a combination of automated and manual procedures. There are four major levels in this hierarchy: Class, Architecture, Topology (fold family) and Homologous superfamily (Orengo et al., 1997). Each level is described below, together with the methods used for defining domain boundaries and assigning structures to a specific family.
Domain Boundary Assignments
All the classification is performed on individual protein domains. To divide multidomain protein structures into their constituent domains, a combination of automatic and manual techniques are used. If a given protein chain has sufficiently high sequence identity and structural similarity (ie. 80% sequence identity, SSAP score >= 80) with a chain that has previously been chopped, the domain boundary assignment is performed automatically by inheriting the boundaries from the other chain (ChopClose). Otherwise, the domain boundaries are assigned manually, based on an analysis of results derived from a range of algorithms which include structure based methods (CATHEDRAL, SSAP, DETECTIVE (Swindells, 1995), PUU (Holm & Sander, 1994), DOMAK (Siddiqui and Barton, 1995)), sequence based methods (Profile HMMs) and relevant literature.
The CATH Hierarchy and Classification
Automated Procedures
If a given domain has sufficiently high sequence and structural similarity (ie. 35% sequence identity, SSAP score >= 80) with a domain that has been previously classified in CATH, the classification is automatically inherited from the other domain. Otherwise, the domain is classified manually, based upon an analysis of the results derived primarily from a range of comparison algorithms CATHEDRAL, HMMs, SSAP scores and relevant literature.
Manual and Automated Procedures Combined
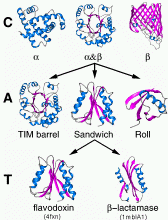
 Class, C-level
Class is determined according to the secondary structure composition and packing within the structure. Three major classes are recognised; mainly-alpha, mainly-beta and alpha-beta. This last class (alpha-beta) includes both alternating alpha/beta structures and alpha+beta structures, as originally defined by Levitt and Chothia (1976). A fourth class is also identified which contains protein domains which have low secondary structure content.
Class, C-level
Class is determined according to the secondary structure composition and packing within the structure. Three major classes are recognised; mainly-alpha, mainly-beta and alpha-beta. This last class (alpha-beta) includes both alternating alpha/beta structures and alpha+beta structures, as originally defined by Levitt and Chothia (1976). A fourth class is also identified which contains protein domains which have low secondary structure content.
This describes the overall shape of the domain structure as determined by the orientations of the secondary structures but ignores the connectivity between the secondary structures. It is currently assigned manually using a simple description of the secondary structure arrangement e.g. barrel or 3-layer sandwich. Reference is made to the literature for well-known architectures (e.g the beta-propellor or alpha four helix bundle).
 Topology (Fold family), T-level
Topology (Fold family), T-level
Structures are grouped according to whether they share the same topology or fold in the core of the domain, that is, if they share the same overall shape and connectivity of the secondary structures in the domain core. Domains in the same fold group may have different structural decorations to the common core.
Some fold groups are very highly populated (Orengo et al. 1994); Orengo & Thornton, 2005) particularly within the mainly-beta 2-layer sandwich architectures and the alpha-beta 3-layer sandwich architectures.
 Homologous Superfamily, H-level
Homologous Superfamily, H-level
This level groups together protein domains which are thought to share a common ancestor and can therefore be described as homologous. Similarities are identified either by high sequence identity or structure comparison using SSAP. Structures are clustered into the same homologous superfamily if they satisfy one of the following criteria:
- Sequence identity >= 35%, overlap >= 60% of larger structure equivalent to smaller.
- SSAP score >= 80.0, sequence identity >= 20%, 60% of larger structure equivalent to smaller.
- SSAP score >= 70.0, 60% of larger structure equivalent to smaller, and domains which have related functions, which is informed by the literature and Pfam protein family database, (Bateman et al., 2004).
- Significant similarity from HMM-sequence searches and HMM-HMM comparisons using SAM (Hughey &Krogh, 1996), HMMER (http://hmmer.wustl.edu) and PRC (http://supfam.org/PRC).
 Sequence Family Levels: (S,O,L,I,D)
Sequence Family Levels: (S,O,L,I,D)
Domains within each H-level are subclustered into sequence families using multi-linkage clustering at the following levels:
| Level | Sequence Identity | Overlap |
|---|---|---|
| S | 35% | 80% |
| O | 60% | 80% |
| L | 95% | 80% |
| I | 100% | 80% |
The D-level acts as a counter within each S100 family and is appended to the classification hierarchy to ensure that every domain in CATH has a unique CATHSOLID classification. The sequence identity and overlap used for clustering are obtained from an implementation of the Needleman-Wunsch algorithm (Needleman & Wunsch, 1970) using a gap penalty of 3. The percentage sequence identity is calculated as (100 * Number Of Identical Residues/Length Of The Shortest Sequence) and the percentage overlap is calculated as (100 * Number Of Aligned Residues/Length Of The Longest Sequence).