Dr Ian Sillitoe
CATH Technical Manager
I am responsible for the technical aspect of CATH. This generally involves maintaining and developing both the front-end interfaces (internal and external web pages and webservices) and back-end code library and databases.
Academic Background
As an undergraduate I studied Chemistry for a 4 year Masters degree at Sheffield University between 1994-1998. I found the topic of organic macromolecular structure and reactions most interesting (specifically protein structure and enzyme catalysis) and this, combined with my dissertation project to create an online resource to teach NMR to undergraduate students, led me to start a Bioinformatics PhD in protein structure/classification here in Prof. Christine Orengo's CATH group at UCL (1998-2002).
After my PhD, I stayed on in the CATH group as a postdoctoral researcher in order to consolidate and extend the work brought up in my PhD and also to help maintain and develop the CATH database (2002-2004). After a couple of years, I decided to have a break from academia and took up a position as the technical director of a commercial web company (2004-2006). A few years later, I decided to have a break from commerce and come back into academia (2006-current)!
Current Research Interests
Protein structure classification and fold recognition
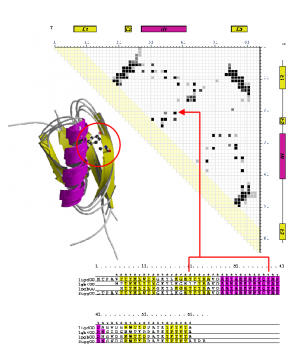
During my PhD, I looked at the classification, analysis and recognition of protein structures using the CATH protein structure classification database. I measured structural similarity by comparing contact maps which are defined as points of contact (<8A) between amino acid residues within a protein. By examining structures that were known to be related, it was possible to identify inter-residue contacts that were highly conserved during the process of evolution.
Part of this project involved deriving a protocol for generating accurate multiple structural alignments and 3D templates based on consensus contact patterns found in these alignments (see figure above). Templates, were generated for all homologous superfamilies in CATH to create a library of unique and identifying 'fingerprint' patterns.
These templates were applied to the recognition of models generated at an early stage of ab initio protein structure prediction. Scanning these early models against a library of templates describing conserved contacts allowed the most likely superfamilies to be indentified. I then wrote an algorithm that performed fold recognition using only a limited set of contacts with the purpose of application to the early stages of experimental NMR structure determination.
The multiple structural alignments were also used to generate a library of hidden markov models (HMMs). These structure-based sequence profiles were thoroughly benchmarked using a strict dataset of remote homologues and added sensitivity to existing sequence scans.
Selected Publications
Sillitoe I, Dibley M, Bray J, Addou S, Orengo C
Protein Sci14p1800-10(2005 Jul)
Pearl F, Todd A, Sillitoe I, Dibley M, Redfern O, Lewis T, Bennett C, Marsden R, Grant A, Lee D, Akpor A, Maibaum M, Harrison A, Dallman T, Reeves G, Diboun I, Addou S, Lise S, Johnston C, Sillero A, Thornton J, Orengo C
Nucleic Acids Res33pD247-51(2005 Jan 1)
Harrison A, Pearl F, Sillitoe I, Slidel T, Mott R, Thornton J, Orengo C
Bioinformatics19p1748-59(2003 Sep 22)
Pearl FM, Bennett CF, Bray JE, Harrison AP, Martin N, Shepherd A, Sillitoe I, Thornton J, Orengo CA
Nucleic Acids Res31p452-5(2003 Jan 1)
de la Cruz X, Sillitoe I, Orengo C
Proteins46p72-84(2002 Jan 1)
Orengo CA, Sillitoe I, Reeves G, Pearl FM
J Struct Biol134p145-65(2001 May-Jun)
Orengo CA, Bray JE, Hubbard T, LoConte L, Sillitoe I
ProteinsSuppl 3p149-70(1999)
Other Interests
I still remember the days when I had time to play piano/keyboard in various soul/blues/jazz bands - now I only get wheeled out for the occasional wedding, bar mitzvah, etc. I also remember the days when I actually used to be quite good at tennis, football and squash. I still play when I get a chance.
I do still go along to ULU Jitsu as often as I can (I was a instructor there from 2005-2007). Any new people that turn up in the lab will probably get hassled mercilessly until they come along to at least one training session (they usually keep coming after that).
Other CATH Team Members
| Person | Description |
|---|---|
| benoit | Former Member In September 2011 I moved to Osaka, Japan, to work as a Post-Doctoral Fellow in Dr Mizuguchi's group at the National Institute of Biomedical Innovation. Research Interests My main research interests include the study of interactions between proteins and other molecules, both at the structural and network levels. |
| clegg | [Andrew with a half-metre sausage, found (and eaten) on holiday in Germany recently] Senior Research Associate, CATH Development A member of the Orengo group since June 2008, I am the technical lead on the FuncNet platform, which brings together an ensemble of protein function analysis tools from various groups around Europe. This work is supported by the EU-funded EMBRACE and ENFIN research networks. |
| cuff | [ Me and my Cat] CATH Manager I am responsible for the general management and manual curation of CATH. Academic Background As a undergraduate, I read for a BSc(Hons) degree in Biomedical Sciences at the University of Durham and then, after deciding I wanted to pursue Bioinformatics research, I took a MSc degree in Information Technology at the University of Teesside (this was all back in the days before MSc courses in Bioinformatics became available!). |
| lee | David Lee [ ] Post Doctoral Research Fellow I work for the Midwest Center for Structural Genomics (MCSG). My responsibilities include selecting protein targets for structure determination, monitoring the success of target selection strategies, and providing homology models of relatives of MCSG structures. |
| lees | Gene3D Since arriving in October 06 I've been doing development of the Gene3D database in collaboration with Corin Yeats. I also maintain the current Gene3D website. I am involved in several collaborations with experimentalists. Recently (June 2009) I have started a new post employed by ENFIN coordinating a chromosome condensation prediction project, with Juan Ranea (Malaga) and the Ellenberg group (EMBL) (amongst others). We are using novel high throughput phenotype data (Ellenberg Group) a… |
| lewis | Tony Lewis [Me in Malaysia] Senior Programmer I was heavily involved in the complete rewrite of the CATH update procedure that culminated in CATH v3.0.0. I am still involved in maintaining and developing CATH in an ongoing consultancy capacity. Academic Background |
| orengo | See departmental staff page |
| perkins | [Me] London Pain Consortium PhD Student I am a member of the London Pain Consortium, an initiative formed in 2002 by a grant from the Wellcome Trust. I am currently moving into the first year proper of my PhD, supervised by Christine and based in the CATH lab, having completed a year of 3 rotations, working on projects with different labs. |
| phil | [Me] Role in CATH I am post doctoral research associate. One of my responsibilities is the target selection database for the Center for Structural Genomics of Infectious Diseases's structural genomics project. Research Interests CSGID applies state-of-the-art high-throughput structural biology technologies to experimentally characterise the three dimensional atomic structure of targeted proteins from pathogens in the NIAID Category A-C priority lists and organisms causing emerging and re-eme… |
| redfern | Dr Oliver Redfern [Posing on the southbank of the Thames] Post-Doctoral Research fellow I work as part of the Midwest Consortium for Structural Genomics, aiding target selection and analysis of the novelty of the protein structures they produce. In parallel, I also develop methods for homology recognition and function prediction from protein structure and sequence. |
| reid | [Me enjoying a traditional Japanese kaiseki meal in a ryokan somewhere outside Kyoto] Me enjoying a traditional Japanese kaiseki meal in a ryokan somewhere outside Kyoto PhD student I am currently nearing the end of my PhD and planning to submit by the end of the year. |
| rentzsch | [Me] Former PhD student I did my PhD in the lab between 2007 and 2012, funded by a EU grant (ENFIN). The ENFIN Network of Excellence aims at close collaboration between experimental and computational groups throughout Europe. I've also worked as a research assistant here. |
| studer | {{ :cathteam:picture.jpg|Me}} {Role in CATH} Description of role in CATH Academic Background Current Research Interests Your research interests go here. Put some pretty pictures in with something like the following: {{ :cathteam:consensus_contact_map_example.png?300 |Example of a consensus structural alignment and contact map }} |
| yeats | Gene3D and BioMiner Gene3D: Design and development, HMM library construction and prediction verification, and web services. Academic Background PhD at the Sanger Institute (2004), supervised by Alex Bateman (Pfam). Thesis: Biological Investigations Through Sequence Analysis. |