GeMMA
As of 08/2012, this page will not be updated until further notice. See further notes below.
Background
Protein domains are the fundamental units of protein sequence evolution. The majority of known proteins contain multiple domains, the particular combination of which can explain protein function.
The domain superfamilies in Gene3D [1] collect homologous domain sequences from all available genomes, extending the 'H' level of the CATH domain classification. Some superfamilies are very large and sequence-diverse and the proteins containing these domains can vary greatly in function.
GeMMA [2] was designed to divide homologous protein and domain sequence families, such as those found in Gene3D and Pfam [3], into functionally conserved clusters. These will help us to study the evolution of sequence, structure, and function at the protein domain level in more detail.
The clustering algorithm
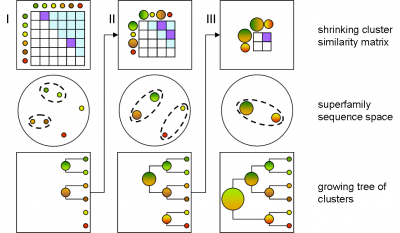
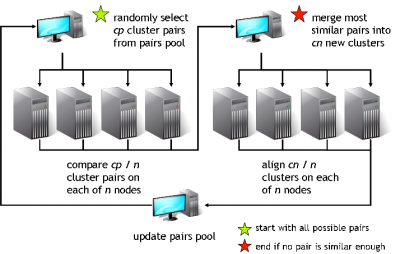
GeMMA performs an all-by-all comparison of a set of sequence clusters and merges the most similar clusters, in an iterative manner. With each iteration, the clusters increase in size, whilst their total number decreases. We currently use COMPASS [4] for profile generation and comparison and MAFFT [5] for multiple sequence alignment (MSA).
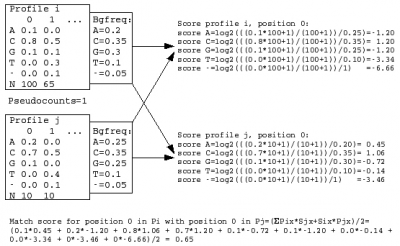
A sequence profile is generated from the MSA of each sequence cluster and compared to all other cluster profiles. Profiles store the observed residue and gap frequencies for each position in a MSA and have been shown to detect remote homology much better than, for example, pairwise sequence comparisons.
Use of domain sequence clusters
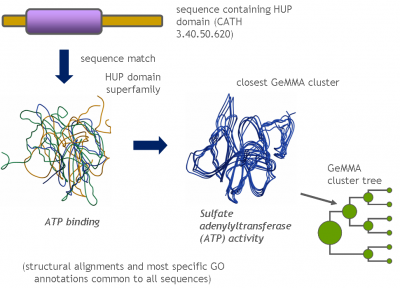
Each GeMMA cluster is much more conserved than its superfamily as a whole; member domains will be highly similar in (partial protein) function and structure. Each cluster can be represented, stored, and queried as a profile hidden Markov model (HMM) or a MSA profile (similar to a Position-Specific Scoring Matrix (PSSM)). This can leverage annotation transfer to unknown sequences, depict family evolution, provide functional categories for comparative genomics studies, and help select targets for Structural genomics initiatives more economically.
Threshold optimization
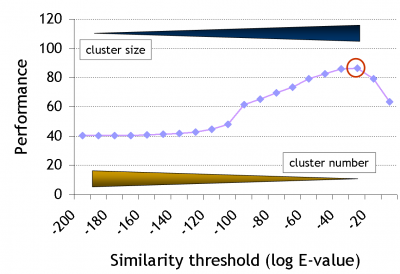
Note that there has been quite some development since the original GeMMA publication, during which the clustering algorithm was retained but the family identification step was entirely revamped. We are moving away from using fixed profile similarity thresholds, towards using available high-quality protein function annotation data, i.e., towards a supervised protocol.
The clustering stops when no remaining cluster pair matches better than a given threshold E (a stochastic 'expectation' value as used in BLAST). Expert-curated domain families from the Structure-Function Linkage Database (SFLD) currently serve as a gold standard to derive E. Good performance in this case means high functional conservation of clusters without over-division of the family.
HPC implementation
For processing large domain families GeMMA was modified to run on the UCL Legion HPC cluster and can be easily set up to run in any environment based on Sun Grid Engine or the Portable Batch System and derivatives such as Torque. However, the iterative nature of the algorithm poses a challenge to both HPC implementation and resources. For most (i.e. the ~2,000 small and medium-sized) superfamilies in Gene3D the current implementation produces clusters within 2-4 weeks.
The following was written in 2010; shortly after we were awarded an Amazon EC2 research grant.
We are currently considering to use resources outside UCL as well, in particular so-called cloud computing. An early adapter bioinformatics project using this technology (there are only few at this point) could benefit both us and the BI community as a whole. It could also help companies providing on-demand HPC and storage resources such as Amazon's EC2 and S3 services to develop solutions tailored more specifically towards BI applications.
If such projects are successful it can be envisioned that bioinformatics groups or even whole universities in the future could focus their resources on infrastructure-on-demand services rather than buying new expensive hardware every two years (enormous sums are currently spent on cooling and powering local HPC facilities).
Different ways of using compute clouds (for different algorithms) should be considered. Options include flow control from scripting languages via API libraries, through middleware as the SGE cloud adapter, and the use of (soon-to-be?) available EC2 AMIs for BI, pre-configured with cluster OSs such as Rocks Clusters.
So-called 'cloud bursting', where external resources are recruited into the same workforce with local nodes and (seemingly) form a single cluster could become a viable tradeoff between availability / bandwidth (local resources) and scalability (grids and clouds) especially for bioinformatics applications, and should thus be tested as well.
References
- Gene3D: merging structure and function for a Thousand genomes.
Lees J, Yeats C, Redfern O, Clegg A, Orengo C
Nucleic Acids Resp(2009 Nov 11) - GeMMA: functional subfamily classification within superfamilies of predicted protein structural domains.
Lee DA, Rentzsch R, Orengo C
Nucleic Acids Res38p720-37(2010 Jan 1) - The Pfam protein families database.
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A
Nucleic Acids Res36pD281-8(2008 Jan) - COMPASS: a tool for comparison of multiple protein alignments with assessment of statistical significance.
Sadreyev R, Grishin N
J Mol Biol326p317-36(2003 Feb 7) - Recent developments in the MAFFT multiple sequence alignment program.
Katoh K, Toh H
Brief Bioinform9p286-98(2008 Jul)