Tutorial on CATH and Gene3D
Introduction
In this practical you will be introduced to the CATH/Gene3D websites and servers that will help you in carrying out an investigation into protein structure and function.
You will begin by looking at the structure of a specific protein of unknown function and methods of assigning function to that family by comparing it with data available on CATH and Gene3D. You will then investigate the structural and functional diversity that can exist within CATH superfamilies by exploring a particularly diverse protein family. Then, you will look at two more clinical challenges, one involving drug design and the other how a pathogenic mutation can effect a proteins structure.
IMPORTANT NOTES
This tutorial involves working though and referring to a number of external websites. It is highly recommended that you click the link with the right hand mouse button and select either open link in new window or open link in new tab so that you don't navigate away from this page.
There are Jmol applets embedded in this tutorial which will allow you to explore a number of different structures. Initially. they will display a simple wireframe model. Please click the gray button next to the applet with your left mouse button to display the structure as required for the tutorial. If for any reason, an applet does not display correctly please refresh your browser.
A Short Introduction to CATH and Gene3D
CATH is a manually-curated hierarchical classification of protein domain structures. The name CATH derives from the initials of the top four levels of the classification - (C)lass, (A)rchitecture, (T)opology and (H)omologous Superfamily.
- Class refers to the secondary structure content (e.g. mainly-alpha, mainly-beta, mixed alpha/beta or 'few secondary structures').
- Architecture refers to the general arrangement of the secondary structures irrespective of connectivity between them (e.g. alpha/beta sandwich).
- Topology, also known as the 'fold' level, takes into account the connectivity of secondary structures in the chain.
- Homologous Superfamily refers to domains that are believed to be related by a common ancestor.
The levels below this, the S, O, L, I and D-levels, are based on increasing levels of sequence identity .
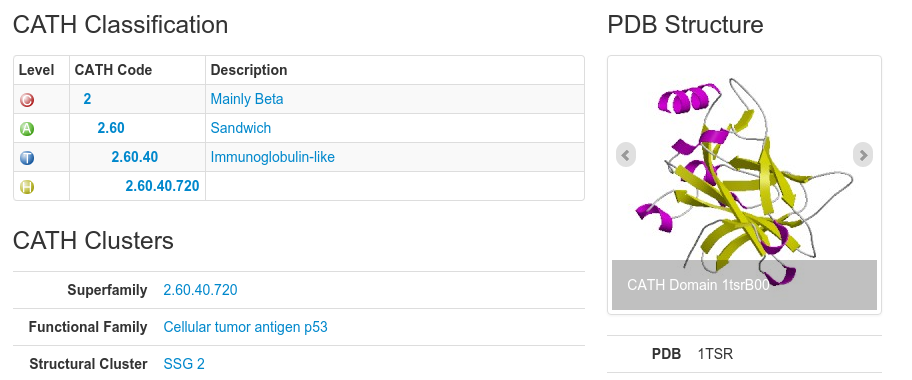
Each level has a CATH code associated with it. Have a look at the following:
In this example, the CATH code for the domain 1tsrB00 is 2.60.40.720. The 2 refers to the class to which the domain belongs (mainly beta), the 2.60 refers to the architecture, the 2.60.40 refers to the actual fold (topology) the domain adopts and 2.60.40.720 is the homologous superfamily code.
The domain code itself (for example 1tsrB00) is broken up as follows: the first 4 letters/numbers make up the domains pdb code, the letter after than refers to the polypeptide chain of the domain you are looking at and the last two numbers refer to the domain number. In the case of a protein chain composing of only 1 domain, the domain number will be 00. Otherwise, the domains will be labeled, 01, 02 and so on.
Gene3D extends the CATH superfamilies to sequenced genomes and the major protein sequence repositories (i.e. UniProt) through the generation of a set of statistical models (hidden Markov models or HMMs) for each superfamily, use of the sophisticated HMM search software HMMER3, and an in-house algorithm called DomainFinder to resolve potential matches into a unified multi-domain architecture (MDA). These predicted sequence domains are presented in Gene3D. Gene3D also merges in many different sources of protein function annotation, ranging from pathway data to active sites, and presents these through a web interface with complex querying abilities.
Assigning function to unknown protein structures
What is the number one question people always have about their protein? What it does! What is the function of the protein you are investigating? Sometimes, we don't know the answer to that, at least not initially. Genomic and metagenomic sequencing projects have provided us with several million protein sequences, around 40% of which will be of unknown function. This number will only increase over time, so we need to develop ways to determine these functions, either by experimentation, or by predicting function using computational techniques.
We are going to explore the function of the protein 2pma.
One of the ways in which the function of an unknown protein can be inferred is by comparing it with the structures of proteins of known function. You can use the CATHEDRAL server to do this. The CATHEDRAL server uses a structural comparison algorithm to compare a protein of interest (otherwise known as the 'query structure') against domains already classified in the CATH database. This means you can try to identify an unknown protein by comparing it with all known structures in CATH.
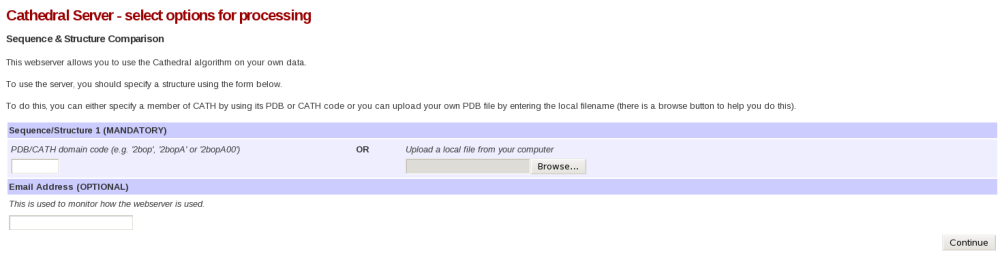
The CATHEDRAL server can be found here. Please click the link. This will take you to a page that looks like this:
Please then input 2pmaA into the PDB/CATH domain code field and press Continue. You have now submitted a CATHEDRAL job on our server. You might find that the job takes a few minutes to complete. If this is the case, please keep refreshing the page every minute or so until the results are displayed.
Alternatively, if the results are taking too long, click the following link: CATHEDRAL results.
The results are sorted by a score calculated by the weighted average of, for example, normalised RMSD, percentage overlap, sequence identity and SSAP score, with those comparisons with the highest scores at the top of the page. The first result is the chain A of 2pma hitting itself, but the second is a different structure (2rspA00). You will notice that the domain IDs on the CATHEDRAL results page are hyperlinked to their entries in CATH.
The box below shows a 3D structural superposition between the two domains 2pmaA01 and 2rspA00 displayed using the program Jmol. What you see initially is a wireframe representation of the superposition, which isn't very clear for this purpose, but if you press the grey button labeled Click here, the two domains will be coloured differently and the wireframe representation will be replaced by a cartoon representation of the structures, making it much easier to compare them. 2pmaA01 is the blue structure and 2rspA00 the red.
<jmol :playground:2pmaa012rspa00.pdb.gz 400 400> jmolButton( “cartoon on; cpk off; wireframe off; select *A; color blue; select *B; color red ”, “Click here” ); </jmol>
You can move the structures around by moving the mouse when within the box while holding down the left hand button, and you can zoom in and out by moving the mouse forward and back while holding down the middle button (or track ball). If you press the right hand button on the mouse when in the box, a menu will pop up; please feel free to explore the structures further by selecting the various options. You can always reset the superposition to its initial state by refreshing your browser.
Question
- Looking at the superposition, are these two domains 2pmaA01 and 2rspA00 similar in structure?
- Look at the CATH entries for both 2pmaA01 and 2rspA00. Which CATH superfamily do they belong to?
Answer
- The superposition suggests that 2pmaA01 and 2rspA00 are very similar in structure.
- They both belong to the superfamily 2.40.70.10.
At the bottom of the page for both entries will be a link to PDBSum ( ). PDBSum pulls in information about a particular structure from a wide range of external resources. Have a look to see if there are any similarities between the two.
). PDBSum pulls in information about a particular structure from a wide range of external resources. Have a look to see if there are any similarities between the two.
Question
- Do the CATH entries and/or other external resources tell you anything about the possible function of our unknown protein?
Answer
- The resources suggest that the 2pma protein is an aspartic protease, due to its close structural similarity to 2rspA00.
Investigating Structural Variation
Most superfamilies are structurally and functionally conserved. However, in some of the most highly populated superfamilies (about 4%), there is a great deal of diversity in both structure and function. Such superfamilies allow us to explore protein evolution and, in particular, how structural changes can result in new functions amongst proteins that are evolutionarily related. In this section, you are going to explore the structure-function relations in one of these, the superfamily of the HUP domains. The CATH code of the HUP superfamily is 3.40.50.620.
The HUP Superfamily in CATH
First of all, you are going to look at the HUP superfamily as it is represented in the CATH database. Please click here to access the CATH website.
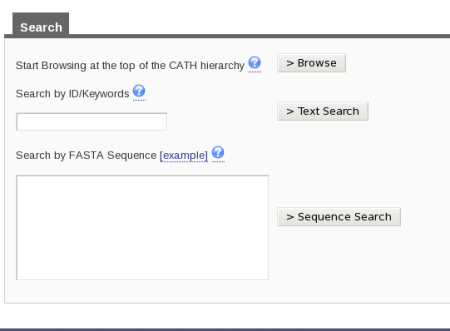
There are two ways in which you can search for a specific superfamily in CATH. At the top right hand side of the home page, there is a search box. You can input 3.40.50.620 into the box and then press the button labeled Search by the side of it. Alternatively, if you look on the home page, you will see the link labeled Search under the title Using CATH and Find my Sequence underneath the title CATH Tools. Click on either of those links and you will be taken to a page that looks like this:
Type in the superfamily code into the search by ID/Keywords box and press the button labeled Text Search.
Either method of searching will take you to a tabulated page of results. Here you will find more information on the superfamily searched for (cathnode), the domains found in that superfamily, the chains associated with the superfamily and also the entire proteins (or pdb entries). By default, you will be directed to the Result Summary tab which displays information for the top hit for each of these four types of data.
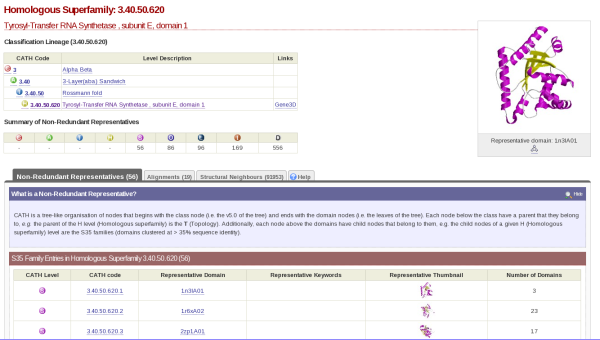
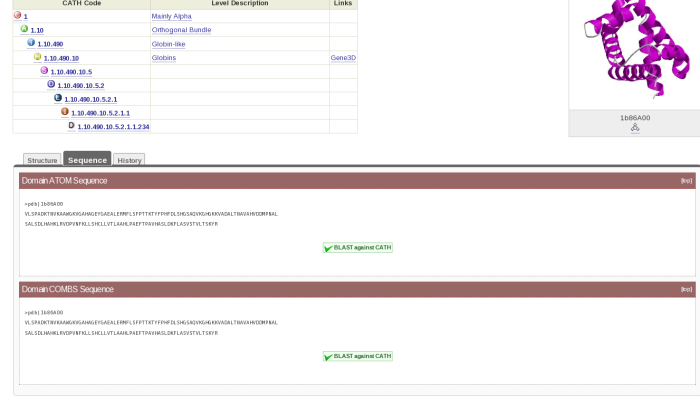
If you click on the cathnodes tab and then click the hyperlinked cathcode displayed, you will be taken to a page that looks like this:
At the top left hand side of the screen there is a table which gives you information on what class, architecture and fold the HUP superfamily (which is described as Tyrosyl-Transfer RNA Synthetase , subunit E, domain 1 in CATH) resides.
Question
- Which well-known fold do the HUP superfamily domains adopt?
Answer
- The Rossmann fold
On the top right hand side, there is the image of a domain representative of the family.
Underneath this is a tabulated display holding different information about the HUP superfamily. By default, you will be shown what is contained by the Non-Redundant Representative tab. Here you will see a list of domains along with hyperlinks to their individual pages and thumbnails of their structures. This list is of the s35 representatives of the superfamily, which means that they each represent a group of structures within that superfamily with a 35% sequence identity; otherwise known as s35 clusters.
Question
- How many s35 clusters make up the HUP superfamily?
- Does the number of s35 clusters suggest anything about the diversity of this superfamily?
Answer
- 69
- Possibly. The number of s35 clusters suggests that there is a significant degree of sequence diversity within the HUP superfamily. However, this does not guarantee structural diversity, as structure is far more conserved throughout evolution than sequence.
The Alignments tab displays groups of domains in the HUP superfamily that have been placed in the same cluster due to being very close in terms of structural similarity.
The HUP superfamily in Gene3D
Now, you are going to use Gene3D to explore the HUP superfamily. Please click here to go to the Gene3D website. From the front page go to the “Get superfamily summary” tab and enter the hups superfamily code 3.40.50.620 and click the “Get Superfamily” button. Which will take you to the a page showing a summary of this superfamily in Gene3D, click here to go directly to this page.
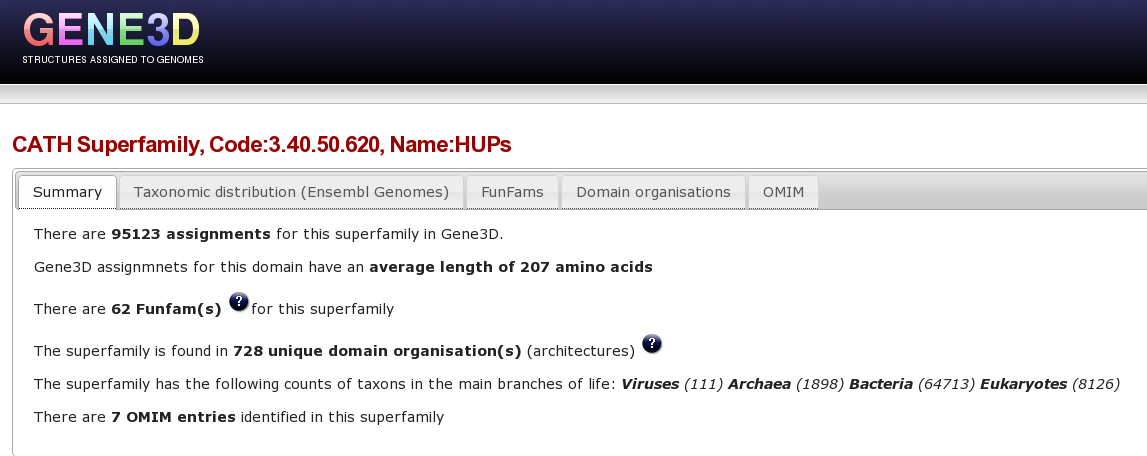
At the top of this Superfamily summary page, there are a number of tabs, the first tab shows a brief summary of stats for this superfamily:
Clicking on each of these items in turn provides different types of information for the superfamily. For example, Clicking on the “domain organisation” tab brings up a page displaying the multiple domain architectures, or MDA's, associated with the HUP superfamily protein domain. It is possible to a column by clicking on it, for example clicking on the “Number of Viruses” column we can see that the HUP superfamily is found in several viral species/strains.
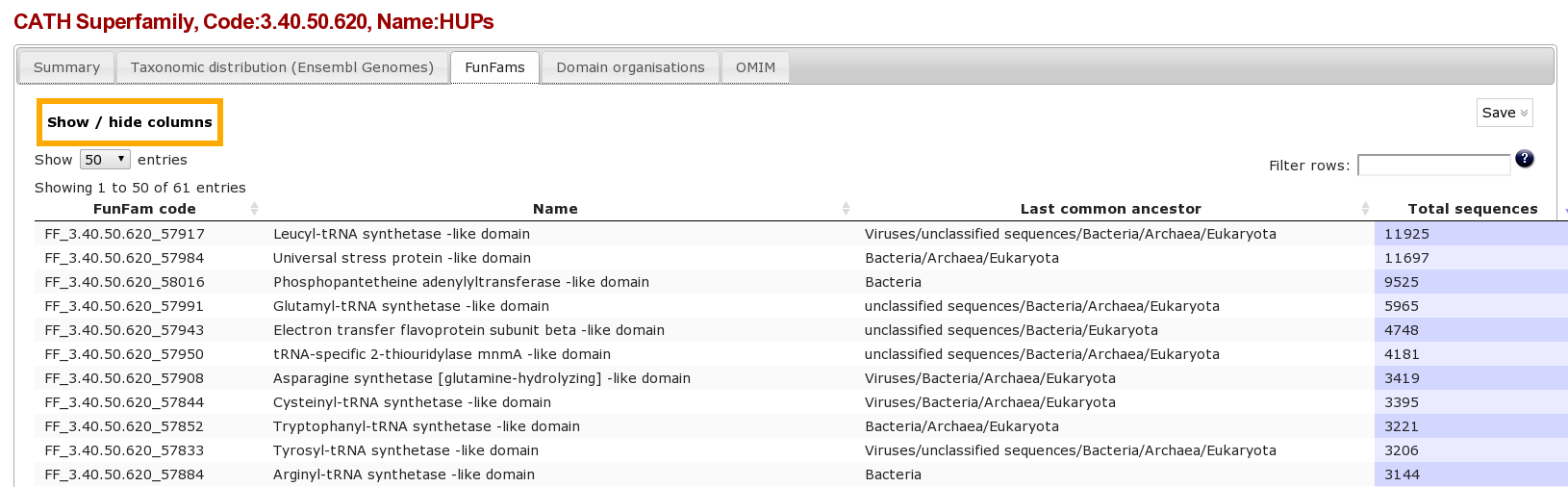
Clicking on the Funfams tab will bring up a page displaying sub-divisions of the CATH superfamily into its functional families (FunFams). These FunFams provide a means of interpreting the sometimes very large and structurally diverse superfamilies at the functional level.
Question
- What is the most highly populated FunFam? (Hint Click on the column header to order the column)
Answer
- FF_3.40.50.620_57917 Leucyl-tRNA synthetase -like domain
Other tabs include the OMIM tab which shows OMIM diseases from a SNP that is located in this superfamily.
Lets retrieve the proteins with mutations in the HUP domain associated with the inherited disorder CITRULLINEMIA, by clicking the “Get Protein” link in the OMIM tab, or click here to go directly to this page.
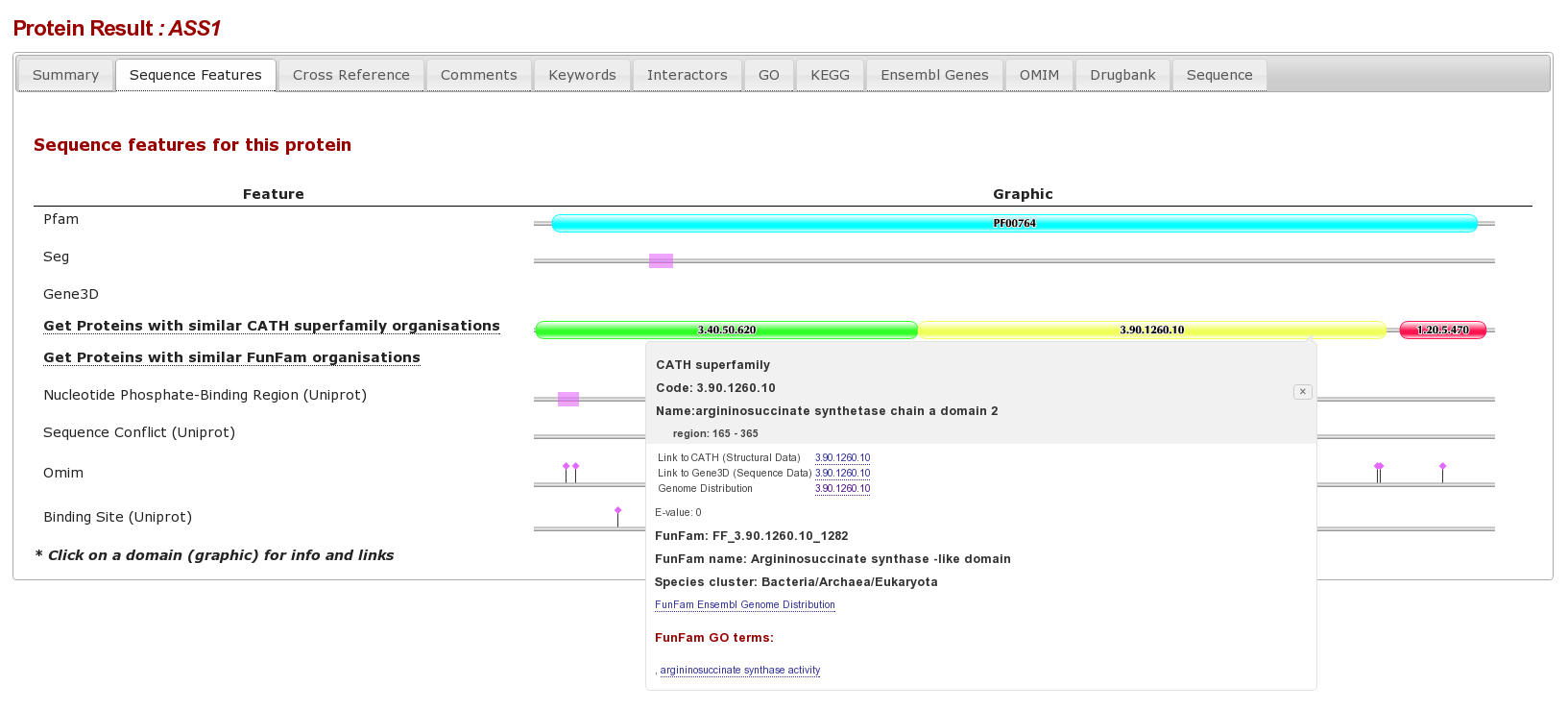
The result page form this is an individual protein sequence, with lots of tabs for different database annotations. Clicking on the “sequence Features tab” shows the various sequence features for this protein, we can see that there are different complimentary interpretations of a domain between PFAM and Gene3D:
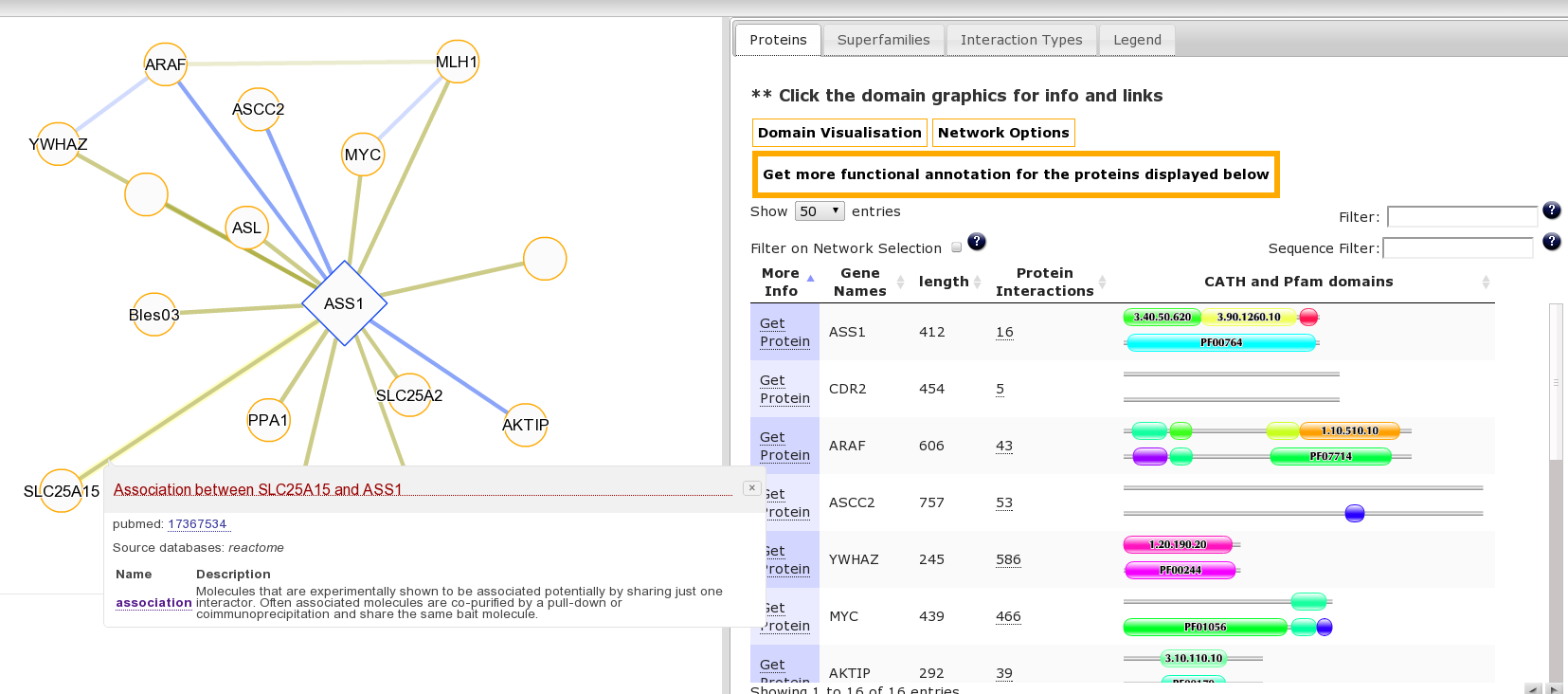
It may be interesting to consider the function of this protein and associated diseases in the context of its interaction partners. This is accessible from the “Summary” Tab for this protein where we can see the protein has multiple protein interaction partners. Clicking the number 16 goes to the protein interaction view or click here to go directly to this page. Clicking on an edge linking two proteins produces a pop-up with details of the source publication supporting the interaction:
Structural Comparison of Two HUP Domains
You are now going to take a closer look at the structural differences that might occur within a diverse CATH superfamily.
In the 3D superposition below, 1r6uB01 is coloured light blue and 1gpmA02 is in pink. Functional residues (namely catalytic residues and ligand binding residues) are highlighted in dark blue and red respectively.
<jmol :tutorials:1r6ub011gpma02.pdb.gz 400 400> jmolButton( “cartoon on; cpk off; wireframe off; select *A; color lightblue; select 310:A, 162:A, 312:A, 309:A, 194:A, 284:A, 313:A, 199:A, 161:A, 163:A, 172:A, 170:A, 173:A, 175:A, 307:A, 200:A, 177:A, 317:A, 339:A, 340:A, 176:A, 337:A, 160:A, 196:A, 338:A, 159:A, 316:A; cartoon off; wireframe 100; color blue; select *B; color pink; select 239:B, 333:B, 400:B, 262:B, 239:B, 334:B, 233:B, 237:B, 335:B, 232:B, 234:B, 265:B, 381:B, 259:B, 315:B, 319:B, 235:B, 240:B, 238:B, 258:B, 260:B; cartoon off; wireframe 100; color red”, “Click here” ); </jmol>
Question
- After investigating the superposition thoroughly, are you able to see any significant differences in the two structures and, if so, what?
Answer
- There are some structural differences evident in the superposition and the functional residues are in different locations in the two structures.
Exploring Drug Design
DNA gyrase (1aj6A00) is a bacterial type II DNA topoisomerase; it plays an essential role in DNA replication and transcription. Novobiocin is an antibiotic that binds to DNA gyrase, inhibiting ATP hydrolysis and therefore its function. A naturally occurring mutation whereby an Arginine is replaced by a Histidine (R136H) confers resistance to this antibiotic due to the creation of a new hydrogen bond between the ligand and the mutant residue at the binding site. The development of new drugs which act on 1aj6 is therefore important.
The 90 kDa heat shock protein (Hsp90) 1a4hA00 belongs to the same superfamily in CATH than 1aj6. Heat shock proteins act as chaperones for a wide range of proteins (referred to as 'client' proteins) involved in, for example, cell cycle regulation. This has lead to the development of potential antitumour drugs that target Hsp90. Geldanamycin is one such compound being researched; it has been shown to interact directly with Hsp90 to promote degradation of 'client' proteins before they become fully active.
You are going to look into the possibility of geldanamycin being a lead molecule for the development of a new drug to act upon DNA gyrase.
First of all, look at the CATH database records of the domains 1a4hA00 and 1aj6A00. Go to the CATH website by clicking here and search for the two domains as explained above.
Question
- What CATH superfamily do the domains 1a4hA00 and 1aj6A00 belong to?
Answer
- They both belong to the superfamily 3.30.565.10
CATH has an in-house structural comparison algorithm called SSAP. SSAP takes two structures and calculates how similar they are in structure, residue-by-residue. Similarity is measured by the SSAP score. this ranges from 0 to 100; a score of 100 would indicate that the two structures were effectively identical. Please click here to go to the SSAP server page. Type in 1aj6A00 as Structure 1 and 1a4hA00 as Structure 2. Press continue. A page will be displayed as follows:
Click on the link and the results should appear. If not, keep refreshing the browser every minute or so until they do. Look at the table of results at the top.
Question
- Looking at the SSAP score, are the two domains similar in structure?
Answer
- The SSAP score is 77.77 The closer the SSAP score is to 100, the closer in structure two domains are. A score of over 77 is indicative of a significant amount of structural similarity.
Please find below the two structures superimposed via SSAP. If you press the gray button, you will see the superposition displayed as a cartoon. The green compound in the middle is Geldanamycin, the ligand that is found bound to 1a4hA00 (hsp90). 1a4hA00 is shown in pink, with the residues that bind to Geldanamycin highlighted in red. 1aj6A00 (DNA gyrase) is shown in light blue with the residues that bind to the antibiotic Novobiocin highlighted in dark blue.
<jmol :playground:1a4ha_1aj6a.pdb.gz 400 400> jmolButton( “cartoon on; cpk off; wireframe off; select *A; color lightblue; select *B; color pink; select hetero; wireframe 100; color green; select 34:A, 36-38:A, 40-41A, 44:A, 79:A, 82-84:A, 88:A, 92-93:A, 98:A, 121-125:A, 136:A, 171:A, 173:A; color blue; select 43:B, 46-47:B, 49-50:B, 73:B, 76-79:B, 81-82:B, 90:B, 94-95:B, 120:B, 165:B, 167:B; color red ”, “Click here” ); </jmol>
Question
- Looking at the position of the ligand binding residues and the position of Geldanamycin, what might you be able to say about the ability of DNA gyrase to bind this ligand?
- What therapetic implications could there be if DNA gyrase could bind easily to Geldanamycin?
Answer
- The superposition shows that the ligand binding residues in hsp90 and DNA gyrase are in very similar positions and both can be seen to be in contact with Geldanamycin. It is therefore very likely that DNA gyrase will be able to bind to Geldanamycin.
- Geldanamycin could be used in place of Novobiocin to treat diseases caused by DNA gyrase.
Sickle Cell Anaemia - How a Single Mutation Can Cause Disease
Sickle cell anaemia is a common inherited genetic disorder. People who suffer from the disease have red blood cells that have an abnormal shape much like that of a sickle. These sickled red blood cells are very fragile and the result is severe anaemia. The disease causes many painful symptoms and can significantly reduce a sufferer's lifespan. The abnormal shape of the cells in individuals with sickle cell anaemia comes from a defective protein within the blood cells themselves. This defective protein is haemoglobin.
The structure of normal haemoglobin is shown below. It is a tetramer, which means that its made up of 4 polypeptide chains. In haemoglobin, these chains are identical and are tightly associated with a non-protein heme group. It is this heme group that is the actual active site of the protein (for more information please click here). Heme is highlighted in green in the Jmol applet below. Please also note the residue highlighted as little balls in the blue chain. You will refer back to this later.
<jmol :playground:1b86.pdb.gz 400 400> jmolButton( “cartoon on; cpk off; wireframe off; select *A; color blue; select *B; color red; select *C; color green; select *D; color yellow; select hetero; wireframe on; cpk 50%; color purple; select 6; cartoon off; cpk 60%; color white”, “Click here” ); </jmol>
So, now you have seen what the normal, or native, structure of haemoglobin looks like, you are going to identify the mutation using sequence information gathered from the CATH website. Both the native, (1b86) and mutated forms (2hbs) of the protein have been classified in CATH. You will compare a sequence alignment of the proteins you are interested in. There are a number of online sequence comparison tools; the one you will be using here is ClustalW2. Please click here to access the ClustalW2 website. If you scroll down the page, you should find a box titled Enter or paste a set of sequences in any supported format. This is where you will cut and paste the sequences for the two proteins.
You now need to retrieve the sequence information for the native and mutant forms of haemoglobin in order to perform the sequence alignment.
Go back to the CATH website by clicking here. Now go to the domain page for 1b86B00 as follows. Type the domain code in the search box and press enter. You will get to a tabulated page. Press the tab titled Domains and then click on the appropriate link. Once you get to the domain record page, click on the tab titled Sequence. You should now be looking at a page which looks like this:
Cut and paste the Domain ATOM Sequence into the box on the ClustalW2 page. Then search for the domain 2hbsB00, find the sequence for that and cut and paste it directly underneath the one for 1b86B00. Press the red button titled Run underneath the box. Wait until the sequence alignment has been completed. When you get to the results page, scroll down until you see the alignment. Have a look at the alignment.
Question
- What residue change can you see in the mutated protein domain 2hbsB00? (HINT: You might find it easier to spot if you press the Show Colors button underneath the alignment).
Answer
- The residue glutamate 6 has been substituted for a valine
So, now you have discovered the mutation in haemoglobin that causes sickle cell anaemia, the next step is to find out how that mutation causes the disease. Understanding how a mutation affects a protein is an important step in developing treatments to combat a disease. Many mutations causes disease by changing the active site, and therefore a vital function, of the protein they are affecting. Now, do you remember those little white balls in the Jmol above? They represent the mutated residue, valine 6.
Question
- Looking at the haemoglobin structure above, does the mutation glu6-val affect the active site of the protein?
- Whereabouts on the protein is the mutation located? Is it buried within the structure or on the surface?
Answer
- No, the mutation is in a different location to the active site of haemoglobin.
- The mutation is on the surface of the protein
So, how does the mutation glu6-val cause sickle cell anaemia? Have a look at the Jmol below. This is the mutated haemoglobin structure 2hbsA00. The mutation, as with the native structure, is highlighted as white balls.
<jmol 2hbs 400 400> jmolButton( “cartoon on; cpk off; wireframe off; select *A; color blue; select *B; color red; select *C; color green; select *D; color yellow; select *E; color purple; select *F; color lightblue; select *G; color pink; select *H; color gray; select 6:H; cartoon off; cpk 60%; color white ”, “Click here” ); </jmol>
Question
- What is the major difference between the native and mutant structures of haemoglobin?
Answer
- There are 2 haemoglobin tetramers joined together for the mutant form of the protein
Valine is a hydrophobic amino acid, which means it doesn't like being surrounded by water. It therefore tries to bond with other hydrophobic residues - in this case a phenylalanine and a leucine from another haemoglobin molecule. This causes the association of 2 individual haemoglobin molecules as seen above.
Long fibres of haemoglobin molecules form as the mutated valine-6 residues just keep adding on more haemoglobin molecules as they try to stabilize their structure. As the fibres form, they cause the shape of the red blood cell to become sickle-shaped. The long fibres push the cell membrane out of shape, causing the characteristic shape of the red blood cells in the disease. These cells can no longer move normally through the blood vessels, so normal delivery of oxygen to the body is interrupted. This is what causes the disease, sickle cell anaemia.